Research in the Baltrus Lab
Our research focuses on understanding the genetic basis for bacterial interactions with other organisms (be they plants, insects, fungi, other bacteria), and on how evolution shapes these interactions. We approach these questions with a variety of tools, from screening for mutants using "toothpicks and agar plates" to experimental evolution to comparative genomics (and pretty much everything in between). A variety of our current research projects are described below.
|
Photo courtesy Betsy Arnold
Photo courtesy Kevin Hockett
|
Endohyphal Bacteria within Endophytic Fungi
We, as part of a shared project with the Arnold and Gallery labs, are currently investigating genetic, genomic, and evolutionary questions for a diverse array of bacteria found living inside of fungi. How are such symbioses established? What benefits do the bacteria receive from fungi? Do bacteria have host ranges? (and many more questions!!!) For more information, please see http://www.endohyphalbacteria.com/ Bacteriocin Genetics and Evolution in Pseudomonas
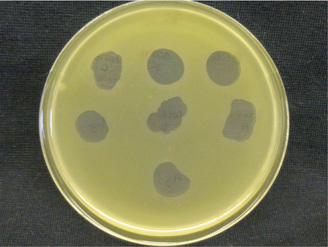
We have discovered that Pseudomonas syringae (and other species) have coopted bacteriophage tails into compounds that can kill closely related strains and species. Even though phage derived bacteriocins of P. syringae are located in the same genomic context as R-type pyocins of P. aeruginosa, these compounds were independently derived from different progenitor phage. Our current research focuses on understanding evolutionary pressures that drive changes to bacteriocins within environmental Pseudomonads. Microbiomes of Invasive Species We and the Dlugosch lab are investigating if and how microbiomes differ for yellow starthistle (and invasive weed in the United States) between its native and invasive ranges. Evolutionary Effects of Horizontal Gene Transfer
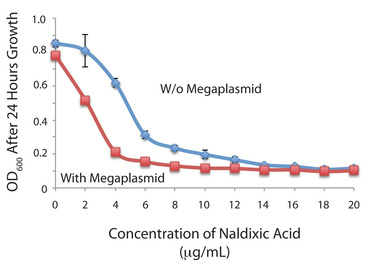
HGT, the movement of genes between individuals, is rampant within bacterial communities. We are investigating the genetic basis of secondary phenotypic changes associated with HGT (for instance acquisition of a 1Mb megaplasmid in Pseudomonas lowers resistance to some antibiotics, left) as well as how such HGT events affect future evolutionary trajectories within bacterial populations. Virulence Evolution in Pseudomonas
We are interested in using comparative genomics and genetics to better understand the origin of pathogenicity in Pseudomonas across host species. Much of our current work focuses on studying phytopathogenicity and entomopathogenicity of Pseudomonas syringae . |